Illumina
Illumina is the current gold standard in short-reads sequencing. Illumina is based upon sequencing-by-synthesis technology and can generate billions of small fragments.
Sequencer Specs

| Platform | Max Number of Cycles | Flow Cell Size | Data Output (reads max) | Data Output (Gb max) |
|---|---|---|---|---|
| MiSeq V2 | 600 Cycle | 1 flow cell with no divisible units | ~10M paired end reads/flow cell | ~4.5-7.5 Gb/flowcell |
| MiSeq V3 | 600 Cycle | 1 flow cell with no divisible units | ~20M paired end reads/flow cell | ~13 Gb/flowcell |
| NextSeq | 300 Cycle | 1 flow cell with no divisible units | ~400M paired end reads/flow cell | ~120 Gb/flowcell |
| HiSeq X | 300 Cycle | 1 flow cell with 8 divisible lanes | ~350M paired end reads/lane | ~106 Gb/lane |
| HiSeq 2500 | 500 Cycle | 1 flow cell with 2 divisible lanes | ~150M paired end reads/lane | ~75 Gb/lane |
| NovaSeq SP | 500 Cycle | 1 flow cell with no divisible units | ~650M paired end reads/flow cell | ~200-350 Gb/flowcell |
| NovaSeq S1 | 300 Cycle | 1 flow cell with no divisible units | ~1.3B paired end reads/flow cell | ~400 Gb/flowcell |
| NovaSeq S2 | 300 Cycle | 1 flow cell with 2 divisible lanes | ~3.3B paired end reads/flow cell | ~1 Tb/flowcell |
| NovaSeq S4 | 300 Cycle | 1 flow cell with 4 divisible lanes | ~8B paired end reads/flow cell | ~2.3 Tb/flowcell. ~600 Gb/lane |
How Illumina Works
Illumina sequencing operates following the below schematic.
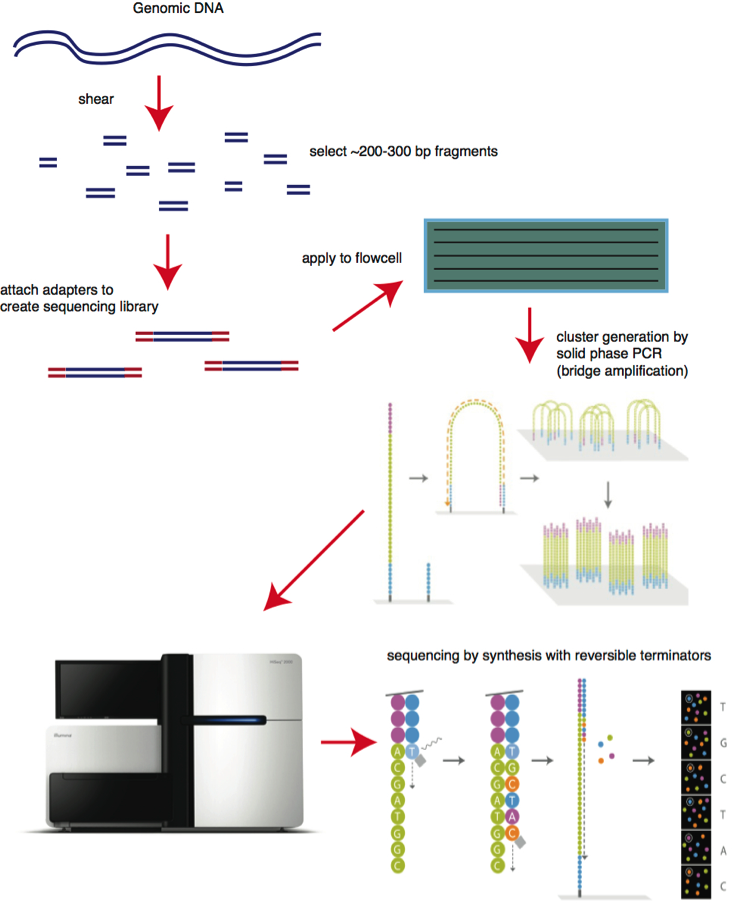
Bridge Amplification
After you have done the appropriate library generation, you bind the library to the flow cell. The flow cell is seeded with oligonucleotides that are complementary to the adapter sequences. Once bound, there are several rounds of amplification. This is done as “bridge amplification” where the fragment arches over like a bridge to the other adapter complementary site and allows the amplification of one molecule into a cluster of molecules- hence the term clustering.
Sequencing by Synthesis
Once the clustering is complete, sequencing by synthesis begins. The flowcell is flooded with fluorescently bound terminating nucleotides. This allows the incorporation of one base to each cluster (via polymerase), and then the laser can scan the flow cell to read the fluorescence and assign each cluster a base. The terminator is then cleaved off, and the “cycle” repeats. Illumina kits are usually sold by cycle, and this is how many times you can add a base in this fashion.
The MiSeq and HiSeq models use a 4 laser chemistry (red, blue, green, yellow) with each base being a different color. The NextSeq and NovaSeq use a 2 laser chemistry (red, green, dual, null) which allows for cost reduction and accelerated scanning.
Library Construct
Once the library is constructed, you will have your insert in the center which is usually 200-400bp in side. On each pole, you will have the Illumina adapter sequence and any applicable indices.

Insert size is important as too small or too large will cause clustering to fail. 200-400bp is the average, with 100bp being the lower limit and 700bp being the upper limit.
Order of sequencing
Order of sequencing is critically important to remember if doing a custom library design. Read 1 from the P5 adapter is sequenced first, followed by Index 1 from the I7 index. If dual indexing is included, then the I5 index is read as Index 2, followed lastly by Read 2 from the P7 adapter. If you are doing single indexing, ONLY I7 WILL BE USED.