PacBio
Pacific Biosciences (PacBio) introduces the concept of long read sequencing. Whereas short-read sequencing has historically been 200-400bp, and some of the longer technologies like Sanger can reach ~1000bp, PacBio has the target average of 10-40Kb. You can target even larger as well, with the PacBio records being >100Kb. The 10-40Kb is usually the target though, as the circularized nature of the read allows for inherent error correction to compensate for the higher error rate of long reads sequencing.
Sequencer Specs
| Platform | Read time | Flow Cell Size | Data Output (reads max) | Data Output (Gb max) |
|---|---|---|---|---|
| RS II | Up to 4 hours | 1 flow cell with up to 16 SMRT cells | ~55,000 reads | ~1 Gb/SMRTcell |
| Sequel | Up to 20 hours | 1 flow cell with up to 16 SMRT cells | ~300,000-500,000 reads | ~8-10 Gb/SMRTcell |
| Sequel II | Up to 30 hours | 1 flow cell with up to 16 SMRT cells | ~3-4M reads | ~40-50 Gb/SMRTcell |
How PacBio works
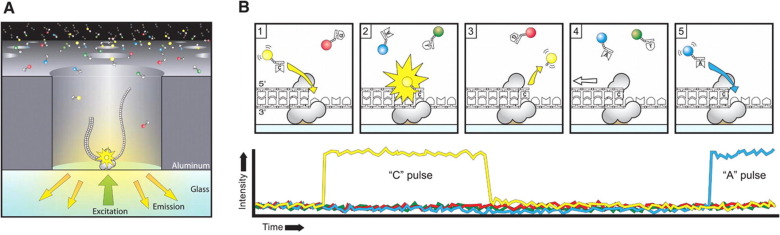
PacBio sequencers are also based on the premise of sequencing by synthesis, but in a different fashion. They utilize Single Molecule Real Time (SMRT) sequencing. For this, they generate their libraries (see below) and then load the flow cell to get a single molecule per zero-mode wavelength (ZMW- essentially a well on the flow cell). The circularized fragment is now sequenced by synthesis, but instead of using terminating nucleotides with laser scanning fluoroscence, PacBio utilized video capture to see basepair fluorescence as it is incoporated.
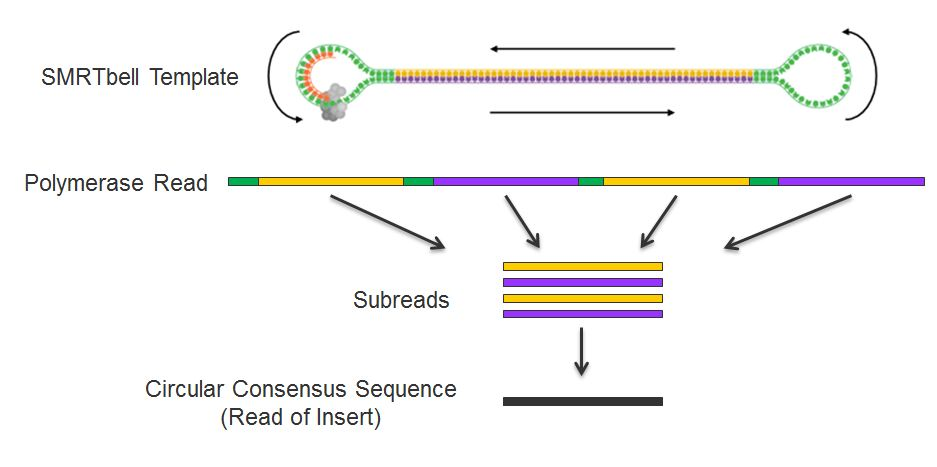
PacBio library construct
PacBio libraries are constructed to be long-reads. To do this, they add SMRT bell adapters to the poles of the double stranded fragment. From there, the fragment in circularized and bound with polymerase. As it is sequenced, the reads can be collapsed by splitting the reads at the known SMRT bell adapter sequences.