Welcome to the Next Generation Sequencing Knowledge Base. Please see each section below for links to each primary sequencing technology currently active. Links are additionally in the sidebar for navigation.
Knowledge Base Categories
Common Terms to know
Reads
The string of A-T-C-G bases that comes from the sequencer. Usually this is combined from thousands -> millions of individual reads into a .fastq or .fasta file.
.fasta file
This is the most basic file for NGS data, it is just the individual strings on bases with each line delineated with a “>” header.
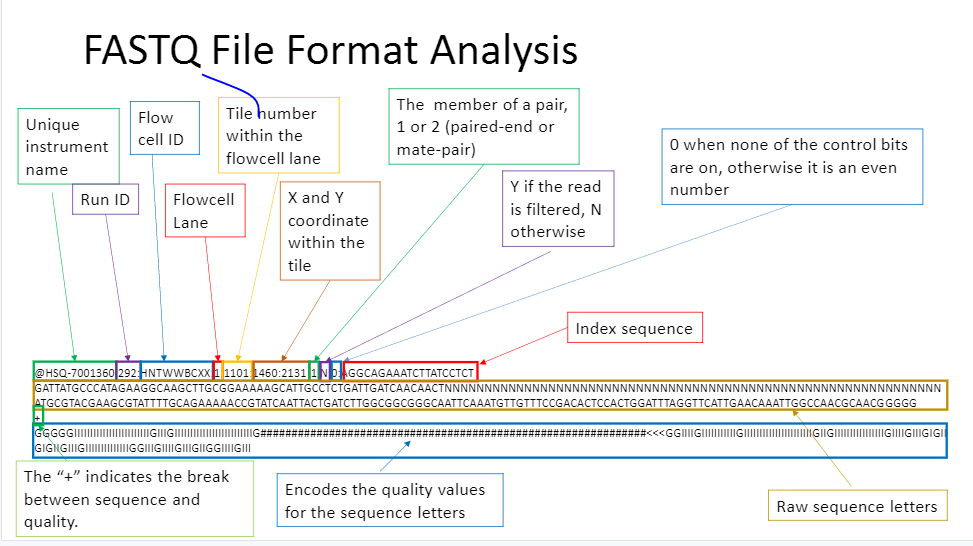
.fastq file
Similar to the .fasta file, but includes additional information. This is more common for sequencer reads as it includes quality information and details on the sequencer usually. Much of it is only relevant the the sequencing provider, but the quality is important to everyone. fastq files are split by the “@” header to define a line, and the “+” indicates the different line for base call and quality encoding.
Coverage
The number of times a base has been covered by sequencing reads. It can be calculated by: Basepairs sequences/target size
So for example you have the human genome (~3Gb) and you have 350M reads of 150bp size
coverage = (350000000*150)/300000000
coverage = 17.5
And this is usually denoted at 17.5X coverage.