Whole Genome Sequencing Library Preparations
Whole genome sequencing (WGS) requires generating sequencer compatible libraries from genomic DNA. This can be done a variety of ways, but in essence is fragmenting the DNA, adding sample indices, and adding the final sequencing adapter poles.
Sonic Fragmentation
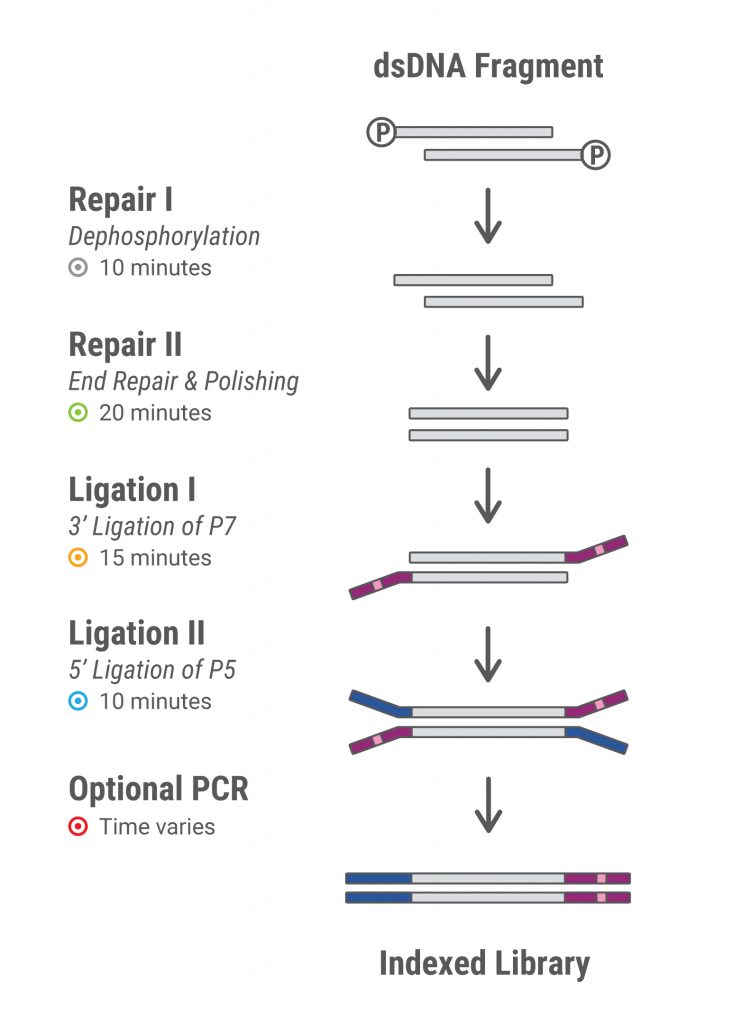
One of the historical methods of preparing the genomic DNA for sequencing is sonic fragmentation. Depending upon your organism of interest, this can require some optimization of the sonication protocol to generate appropriate size fragments. The target is the ~200-400bp insert of random gDNA fragments.
The most common library prep utilizing this workflow is the New England Biolabs (NEB) kits. In brief, you start with your fragmented DNA, perform end repair and polyA tailing for initial adapter ligation. Once the adapter scaffold is ligated, you can perform indexing PCR to add the applicable sample indices and adapter poles to the fragments. This allows you to properly bind each fragment to the flow cell, as well as delineate each sample based upon the index.

PCR Free
PCR free library preps are very similar to the standard fragmentation workflows excpet they do not incorporate any PCR steps (what a shocker- I know, its only called “PCR-free”). In essence the workflow includes end repair, then direct full adapter ligations with indices. The downside is that this usually requires higher input materials that may cost most due to higher reagent burn, but you do get removal of library PCR biases.
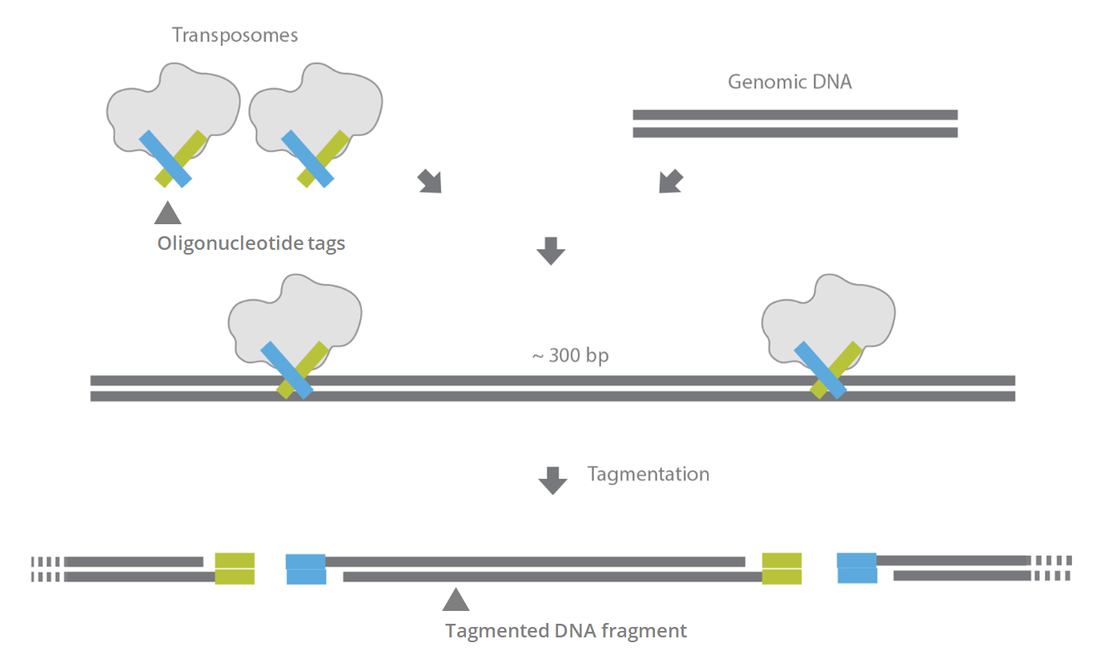
Tagmentation
As an alternative to sonic fragmentation, a common current workflow is using “tagmentation”. Tagmentation is the use of a hyperactive transposase (Tn5) to insert into the gDNA and fragment it- during which it directly incorporates the base adapter. This allows for quick fragment generation that then only needs indexing PCR to add the indices and adapter poles. This method can often go to lower limits of input gDNA, but does require a more controlled gDNA concentration. The input concentration and tagmentation time determines the size of the fragment, so it must follow proper protocols to achieve the target of ~200-400bp insert. The traditional kit for this is the Nextera XT workflow, which has been upgraded into the Nextera Flex by Illumina.